STUDY OF MACHINE LEARNING ALGORITHMS TO ASSIST IN THE PREDICTION OF NEW MATERIALS
Macedo, B. H. D. - Graduating in Engineering Physics - Latin American Institute of Life and Nature Sciences (ILACVN) - Federal University of Latin American Integration - UNILA, North Region, Foz do Iguaçu - PR, Paraná, Brazil, Email: brunohdmacedo@gmail.com
Zalewski, W. - PhD in Computer Science - Latin American Institute of Technology, Infrastructure and Territory (ILATIT) - Federal University of Latin American Integration - UNILA, PTI, Bloco 6, Espaço 1, Foz do Iguaçu, Paraná, Brazil, Email: willian.zalewski@unila.edu.br

Abstract
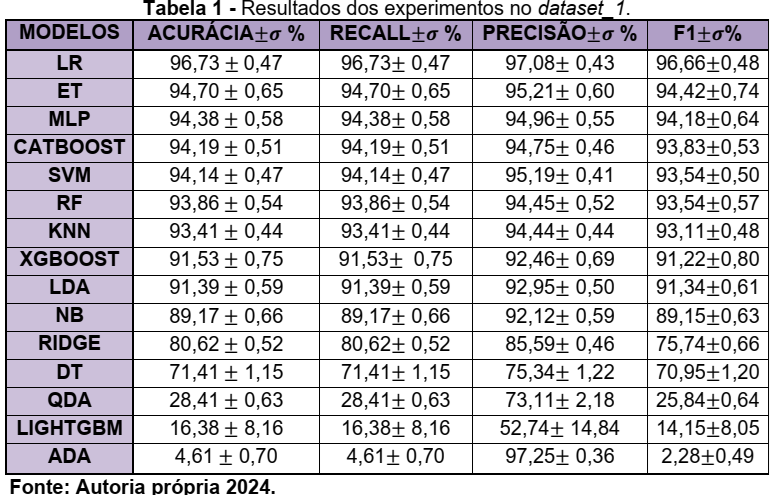
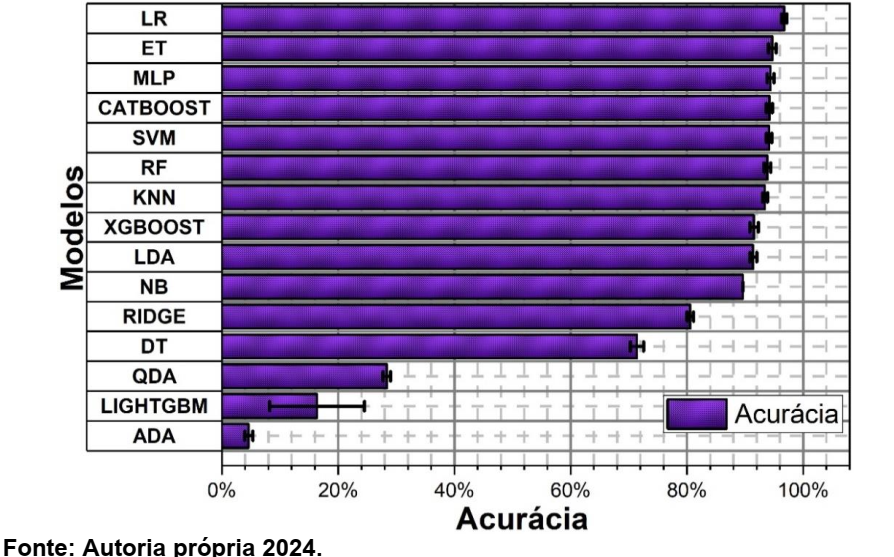
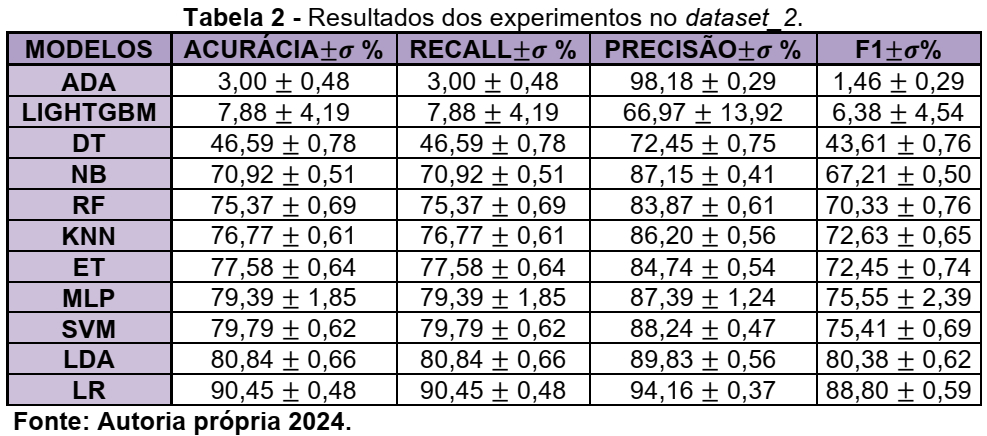
In recent years, materials development has been essential for technological advances in areas such as electronics, aerospace, and automotive. Discoveries such as semiconductors and light and resistant alloys have driven innovations (Schleder & Fazzio, 2021). However, traditional discovery methods, based on empirical experimentation, are slow and expensive. Advances in computational capacity and the creation of large databases have opened up new opportunities for materials science (G1, 2024). The integration of simulations and experimental data with Machine Learning techniques has revolutionized the field, enabling the analysis of large volumes of data, identification of patterns, and prediction of properties (Schleder et al., 2019; Wang et al., 2020). This, combined with high-precision simulations and laboratory experiments, has enabled significant optimizations in areas such as energy and biomaterials (Ramprasad et al., 2017). Repositories such as the American Mineralogist Crystal Structure Database, Crystallography Open Database, ICSD, and RRUFF are essential for crystalline data, while the CCDC is used for non-crystalline materials (Pagliarini, 2022)[1]. This research aims to evaluate the use of Machine Learning algorithms on data from the RRUFF dataset, which contains 1704 classes and 8950 spectra, divided into two sets: Dataset_1 with 192 classes and 5292 spectra, and Dataset_2 with 1332 classes and 8578 spectra. In the work of (SANG et al., 2021), a 1D CNN was developed for mineral classification from Raman spectra, with superior performance to classical models such as KNN, Decision Tree, Random Forest, and SVM. Comparing the results, the CNN obtained 98.43% accuracy, surpassing KNN (k=1~96.76% and k=5~93.76%), Decision Tree (77.76%), Random Forest (74.60%) and PCA+SVM (linear~97.72% and rbf~86.67%). Initially, we downloaded and preprocessed the data using the python programming language to construct dataset_1 and the baseline as proposed by Dourado Macedo, B. H., & Zalewski, W. (2024)[3]. The BayesSearchCV function from the scikitoptimize library was applied to explore the best combination of parameters. The models were evaluated using the train_test_split strategy. To do this, we used a for loop with range(30) to divide the data into 4 partitions X_train, X_test, y_test and y_train for each loop excursion. Thus, in each repetition, different data are selected for each partition, thus minimizing possible bias in the data. The parameters used for this evaluation were: X_train, X_test, y_train, y_test = train_test_split(X_, y, train_size=0.7, test_size=0.3, random_state=i, shuffle=True, stratify=y) and cv_inner = StratifiedKFold(n_splits=2, shuffle=True, random_state=1) to optimize the parameters in each repetition.
Load Spectrum
def load_spectrum_file(dir_path, max_subfolders=None, max_files_per_subdir=None, max_rows=None):
"""Carrega arquivos de espectro e associa rótulos numéricos às subpastas."""
data, labels = [], []
subfolder_labels = {}
label_encoder = LabelEncoder()
# Contadores para controlar a quantidade de subpastas e arquivos lidos
subfolder_count = 0
for root, dirs, files in os.walk(dir_path):
# Limitar a quantidade de subpastas
if max_subfolders is not None and subfolder_count >= max_subfolders:
break
for dir_name in dirs:
subfolder_path = os.path.join(root, dir_name)
subfolder_labels[subfolder_path] = dir_name
subfolder_count += 1
# Limitar a quantidade de subpastas
if max_subfolders is not None and subfolder_count > max_subfolders:
break
file_count = 0
for filename in os.listdir(subfolder_path):
if filename.endswith('.csv'):
if max_files_per_subdir is not None and file_count >= max_files_per_subdir:
break
file_path = os.path.join(subfolder_path, filename)
try:
df = pd.read_csv(file_path, delimiter=',', nrows=max_rows)
y = df.iloc[:, 1].values # Pega os valores da segunda coluna
# Adiciona o rótulo da subpasta à lista de labels
labels.append(subfolder_labels[subfolder_path])
data.append(y)
file_count += 1
except Exception as e:
print(f"Error reading file {file_path}: {e}")
# Transforma os rótulos das subpastas em números
labels = label_encoder.fit_transform(labels)
return np.array(data), np.array(labels), label_encoder.classes_
# Exemplo de uso
print("Loading data...")
data, labels, class_names = load_spectrum_file(
'Data_csv/',
max_subfolders=None,
max_files_per_subdir=None,
max_rows=None
)
print(" ")
print("-----------------------------------")
print(" ")
print(f"Data shape: {data.shape}")
print(f"Labels shape: {labels.shape}")
print(f"Class names: {class_names}")
print(" ")
print("-----------------------------------")
print(" ")
X_=data
#print(X_)
y=labels
#X_ = np.nan_to_num(data) # Substitui NaN e inf por zero
#print(X_)
Experiment
# Definição do experimento
def experiment(model_name, model, params, X_train, X_test, y_train, y_test, repet):
# Caminho da pasta para salvar o arquivo (cria uma pasta para cada modelo)
model_dir = os.path.join('Results/', model_name)
# Verifica se o diretório existe, se não, cria o diretório
if not os.path.exists(model_dir):
os.makedirs(model_dir)
# Nome do arquivo para salvar os resultados
results_file = os.path.join(model_dir, f'{model_name}_repetição_{repet}.csv')
# configure the cross-validation procedure
cv_inner = StratifiedKFold(n_splits=2, shuffle=True, random_state=1)
# define search
search = BayesSearchCV(model, params, scoring='accuracy', cv=cv_inner, n_iter=3, refit=True, random_state=1, n_jobs=1)
# execute search
result = search.fit(X_train, y_train)
# get the best performing model fit on the whole training set
best_model = result.best_estimator_
# evaluate model on the hold out dataset
yhat = best_model.predict(X_test)
# evaluate the model
acc = accuracy_score(y_test, yhat)
prec = precision_score(y_test, yhat, average='weighted', zero_division=1)
rec = recall_score(y_test, yhat, average='weighted')
f1 = f1_score(y_test, yhat, average='weighted')
mcc = matthews_corrcoef(y_test, yhat)
# store the result
results = [[model_name, acc, rec, prec, f1, result.best_score_, result.best_params_]]
# report progress
print(f"Repetição {repet}: {model_name} > acc={acc:.3f}, est={result.best_score_:.3f}, cfg={result.best_params_}")
# save results to CSV, each repetition in a new file
df = pd.DataFrame(results, columns=['model', 'acc', 'rec', 'prec', 'f1', 'best_score', 'best_params'])
df.to_csv(results_file, index=False)
Code Classics Model
model_params = {
#00 Logistic Regression
'lr': {'model': LogisticRegression(),
'params': {
'C': Real(1e-4, 1e4, prior='log-uniform'),
'fit_intercept': Categorical([True, False]),
'solver': Categorical(['newton-cg', 'lbfgs', 'liblinear', 'sag', 'saga']),
'max_iter': [500],
'random_state': [1],
'n_jobs': [1] # Limita para 1 thread
}},
#01 K-Nearest Neighbors
'knn': {'model': KNeighborsClassifier(),
'params': {
'n_neighbors': Integer(1, 50),
'weights': Categorical(['uniform', 'distance']),
'algorithm': Categorical(['auto', 'ball_tree', 'kd_tree', 'brute']),
'p': Integer(1, 5),
'n_jobs': [1] # Limita para 1 thread
}},
#02 Gaussian Naive Bayes
'nb': {'model': GaussianNB(),
'params': {
'var_smoothing': Real(1e-10, 1e-1, prior='log-uniform')}},
#03 Decision Tree
'dt': {'model': DecisionTreeClassifier(),
'params': {
'criterion': Categorical(['gini', 'entropy']),
'splitter': Categorical(['best', 'random']),
'max_depth': Integer(3, 30),
'min_samples_split': Integer(2, 10),
'min_samples_leaf': Integer(1, 10),
'max_features': Real(0.1, 1.0, prior='uniform'),
'random_state': [1]}},
#04 Support Vector Machine
'svm': {'model': SVC(),
'params': {
'C': Real(2**-5, 2**5, prior='log-uniform'),
'kernel': Categorical(['linear', 'poly', 'rbf', 'sigmoid']),
'degree': Integer(2, 5), # Relevante para o kernel 'poly'
'coef0': Real(0, 1), # Relevante para os kernels 'poly' e 'sigmoid'
'gamma': Real(2**-9, 2**1, prior='log-uniform'),
'random_state': [1]}},
##05 Gaussian Process Classifier
'gpc': {'model': GaussianProcessClassifier(),
'params': {
'optimizer': Categorical(['fmin_l_bfgs_b', None]),
'n_restarts_optimizer': Integer(0, 10),
'max_iter_predict': [500],
'random_state': [1]}},
##06 Multi-Layer Perceptron
'mlp': {'model': MLPClassifier(),
'params': {
'hidden_layer_sizes': Integer(10,100),
'activation': Categorical(['logistic', 'tanh', 'relu']),
'solver': Categorical(['sgd', 'adam']),
'max_iter': [5000],
'random_state': [1]}},
#07 Ridge Classifier
'ridge': {'model': RidgeClassifier(),
'params': {
'alpha': Real(1e-4, 1e4, prior='log-uniform'),
'fit_intercept': Categorical([True, False]),
'solver': Categorical(['auto', 'svd', 'cholesky', 'lsqr', 'sparse_cg', 'sag', 'saga']),
'random_state': [1],
'n_jobs': [1] # Limita para 1 thread
}},
#08 Random Forest
'rf': {'model': RandomForestClassifier(),
'params': {
'n_estimators': Integer(10, 500),
'criterion': Categorical(['gini', 'entropy']),
'max_depth': Integer(3, 30),
'random_state': [1],
'n_jobs': [1] # Limita para 1 thread
}},
##09 Quadratic Discriminant Analysis
'qda': {'model': QuadraticDiscriminantAnalysis(),
'params': {
'reg_param': Real(0, 1, prior='uniform'),
'store_covariance': Categorical([True, False]),
'tol': Real(1e-5, 1e-1, prior='log-uniform')}},
#10 AdaBoost
'ada': {'model': AdaBoostClassifier(),
'params': {
'n_estimators': Integer(10, 500),
'learning_rate': Real(1e-3, 1, prior='log-uniform'),
'algorithm': Categorical(['SAMME']),
'random_state': [1]}},
##11 Linear Discriminant Analysis
'lda': {'model': LinearDiscriminantAnalysis(),
'params': {
'solver': Categorical(['lsqr', 'eigen', 'auto']),
'shrinkage': Real(0, 1, prior='uniform'),
'tol': Real(1e-6, 1e-4, prior='log-uniform')}},
#12 Extra Trees Classifier
'et': {'model': ExtraTreesClassifier(),
'params': {
'n_estimators': Integer(10, 500),
'criterion': Categorical(['gini', 'entropy']),
'max_depth': Integer(3, 30),
'n_jobs': [1] # Limita para 1 thread
}},
#13 XGBoost
'xgboost': {'model': XGBClassifier(),
'params': {
'learning_rate': Real(0.01, 0.3, prior='uniform'),
'n_estimators': Integer(50, 500),
'max_depth': Integer(3, 10),
'gamma': Real(0, 1, prior='uniform'),
'n_jobs': [1] # Limita para 1 thread
}},
#14 LightGBM
'lightgbm': {'model': LGBMClassifier(verbose=-1),
'params': {
'learning_rate': Real(1e-3, 1, prior='log-uniform'),
'n_estimators': Integer(10, 500),
'num_leaves': Integer(2, 100),
'max_depth': Integer(3, 10),
'n_jobs': [1] # Limita para 1 thread
}},
#15 CatBoost
'catboost': {'model': CatBoostClassifier(verbose=0),
'params': {
'learning_rate': Real(1e-3, 1, prior='log-uniform'),
'iterations': Integer(10, 500),
'depth': Integer(3, 10),
'l2_leaf_reg': Real(1, 10, prior='uniform'),
'border_count': Integer(1, 255),
'bagging_temperature': Real(0, 1, prior='uniform'),
'random_strength': Real(1e-9, 10, prior='log-uniform'),
'thread_count': [1] # Limita para 1 thread
}}
}
Keywords
Ciência dos Materiais, Machine Learning, Aprendizado de Máquina, Raman.